
Research activities Van Zon lab
The central question of developmental biology is how a single fertilized egg develops into a complex multicellular organism.
Even though in the last decades many genes that regulate development have been discovered, it is still an unresolved question how these genes work together to allow cells to sense their environment, communicate with other cells and use this information to make developmental decisions. The observed robustness of development is particularly intriguing given that the underlying processes, such as gene expression and signal transduction, are often highly stochastic. To understand how the collective action of networks of interacting genes drives development in a manner that is robust to stochastic fluctuations, it is increasingly necessary to study development biology in a quantitative manner.
We study these questions in the nematode Caenorhabditis elegans, a small roundworm that has become an important model system for studying development. C. elegans shares many of the molecular mechanisms of development with higher organisms, such as humans. At the same time, it has a simple body plan, a short life cycle and is fully transparent, making it ideally suited for a quantitative, physics-inspired approach. We address these questions using novel experimental techniques, such as single molecule Fluorescence In Situ Hybridization (smFISH), quantitative timelapse microscopy and microfabrication, in combination with quantitative data analysis and mathematical modeling.
smFISH is a novel technique to visualize individual mRNA molecules in fixed, i.e. dead, animals, allowing us to quantify the exact expression levels of any gene in individual cells of C. elegans animals with single molecule resolution. In addition, we are developing an experimental setup that uses very small microfabricated structures and timelapse microscopy to study development quantitatively in live C. elegans animals. Together, these two techniques allow us to quantify the dynamics of development in unprecedented detail.
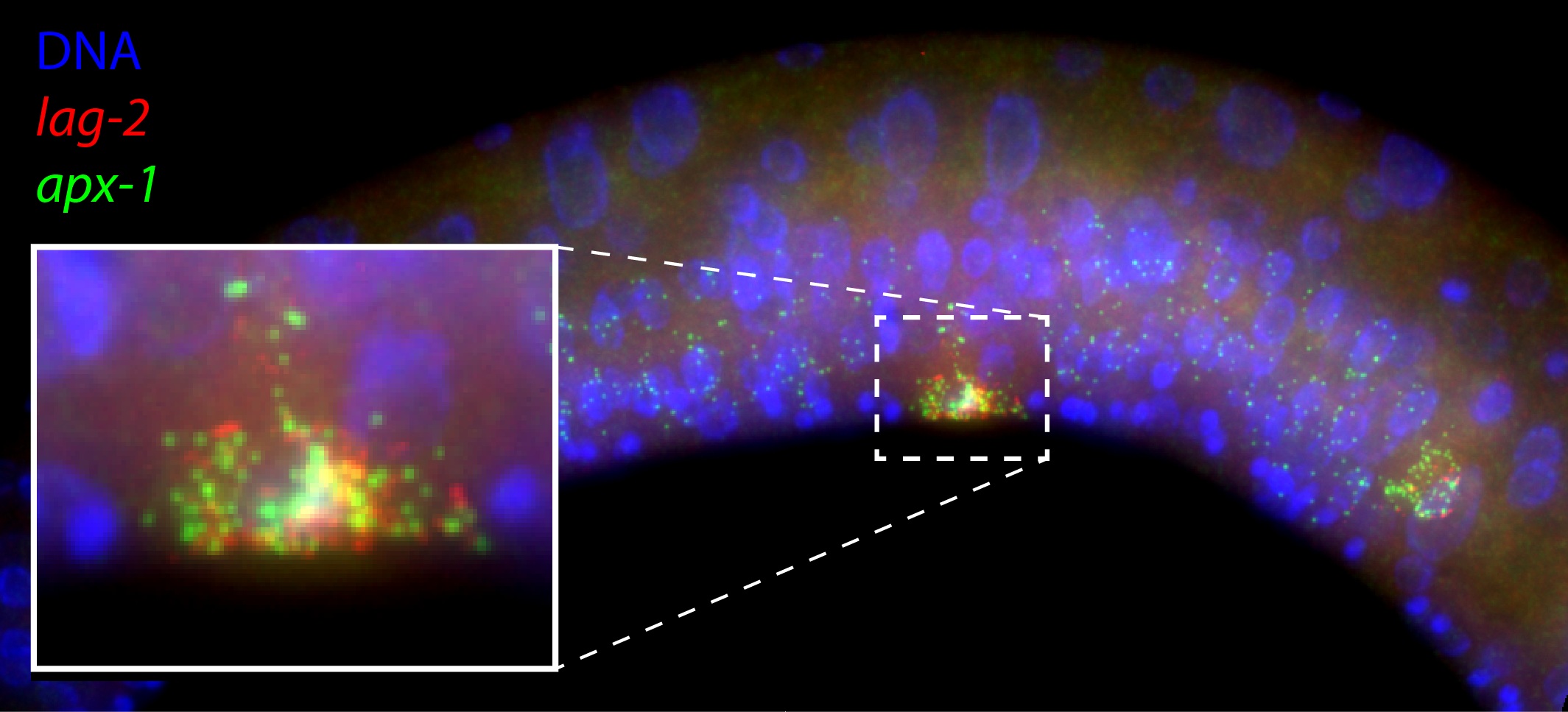
Diffraction-limited single molecule FISH spots in the mid body of a C. elegans larva. Each spot corresponds to a single mRNA molecule of the gene lag-2 (red) or apx-1 (green). Also shown is DNA in the nucleus (blue).
A single growing C. elegans larva locked up in a 200×200 micrometer microchamber. The animal is followed for 12 hours after it hatches from its egg.
Timelapse movie of asymmetric nuclear localization of POP-1::GFP for two rounds of cell division during C. elegans vulva development. The interval between consecutive frames is 10 mins.

