Research activities Biochemical Networks

Cellular Information Transmission
Biochemical networks are the information-processing devices of the living cell. They can perform a variety of computational tasks akin to electronic circuits. Yet, their design principles are markedly different: in biochemical networks the computations are performed by biomolecules such as proteins and DNA that chemically and physically interact with one another. We aim to derive the optimal design that maximizes information transmission given the available cellular resources protein copies, time, and energy. To this end, we use techniques and concepts from statistical physics, stochastic thermodynamics, and information theory.
A list of selected publications can be found here

Circadian Rhythms
One of the most fascinating time devices in biology are circadian clocks, which are found in organisms ranging from cyanobacteria to mammals. These clocks are biochemical oscillators that allow organisms to coordinate their metabolic and behavioural activities with the 24-hour cycle of day and night. Our group employs stochastic simulations and analytical theory to elucidate their design principles and to reveal why and when it becomes beneficial to know the time.
A list of selected publications can be found here
Force Generation By Cytoskeletal Systems
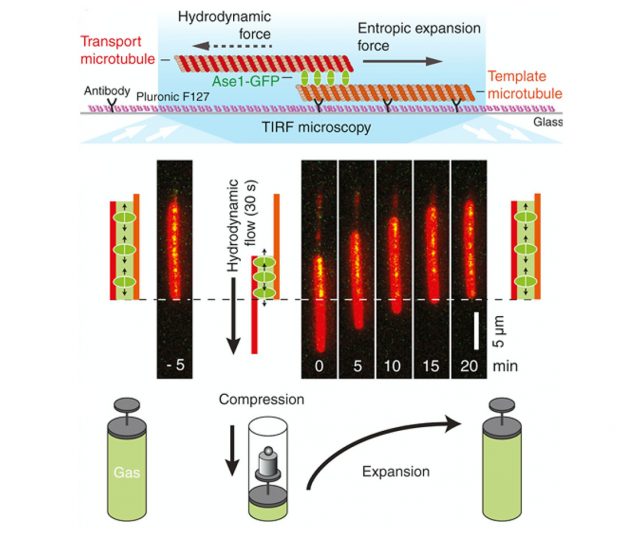
Force generation is of critical importance to living cells. It allows them to move, change shape, and grow and divide. The forces are typically generated via cytoskeletal structures, which consist of filaments that are crosslinked by motor proteins and passive non-motor proteins. Force generation is typically attributed to motor proteins, which consume chemical fuel to do work. Yet, in recent years we have shown in close collaboration with experimental groups that passive crosslinkers can generate forces too, via their condensation to the overlap between the filaments or via the entropy associated with their diffusion within the overlap region. In our group we use statistical mechanics to study not only the condensation and entropic expansion forces, but also the friction forces that oppose these driving forces.
A list of selected publications can be found here
Development New Computational Techniques
GFRD
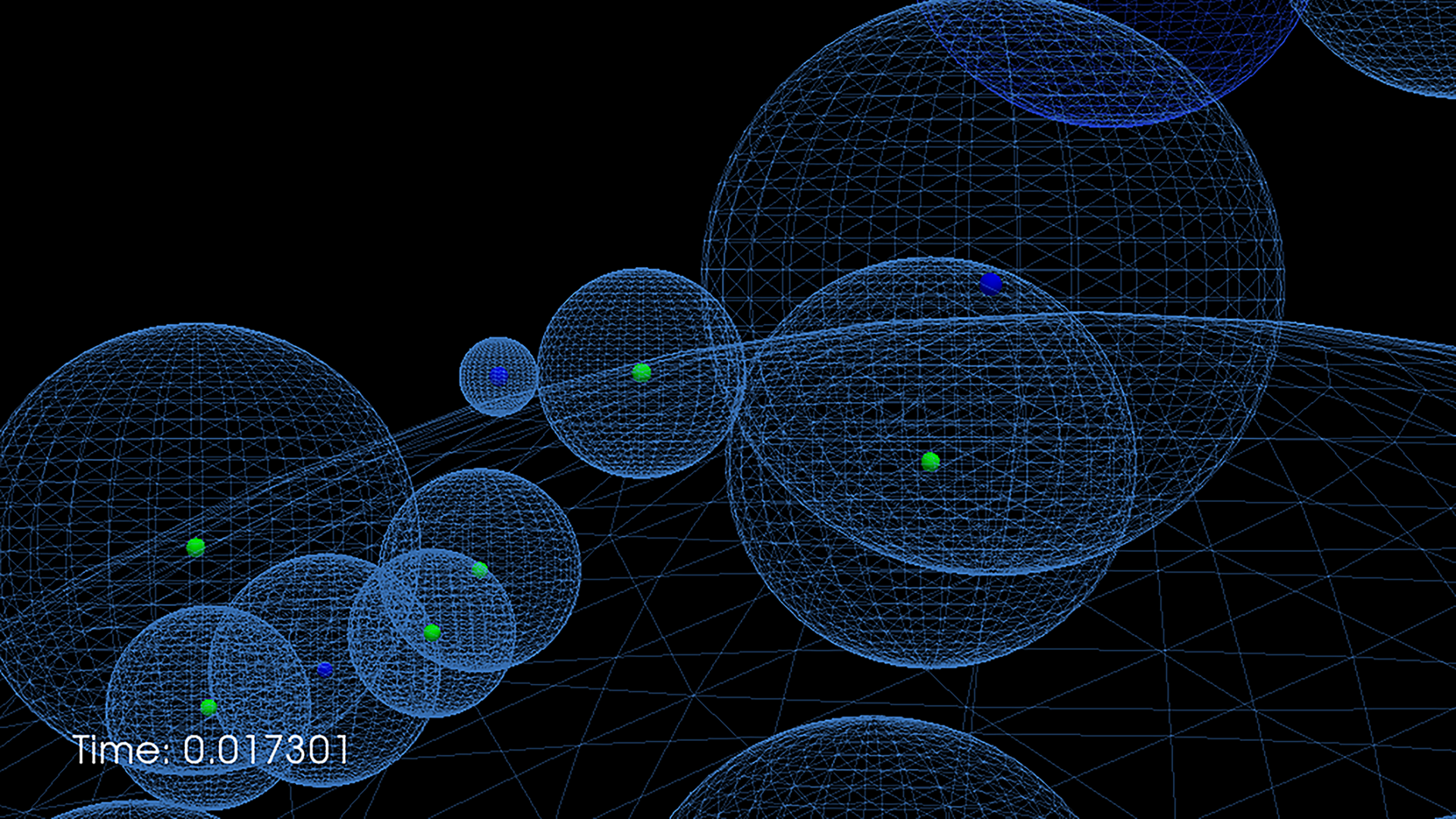
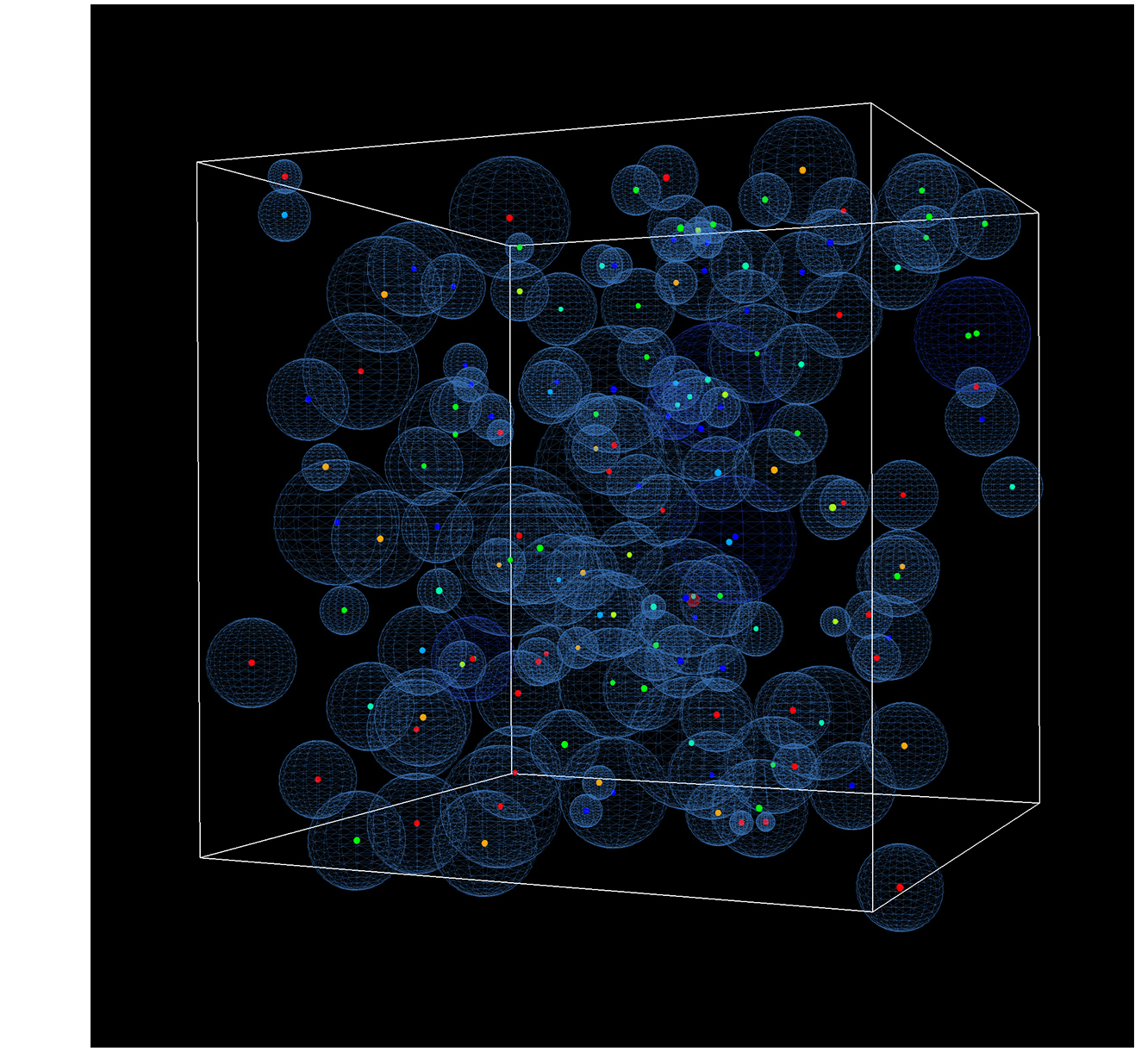
Green’s Function Reaction Dynamics (GFRD) is an exact event-driven algorithm that makes it possible to simulate reaction-diffusion systems at the particle level. At biologically relevant concentrations it is about 4 – 6 orders of magnitude faster than brute force Brownian Dynamics. In recent years we have extended GFRD to include passive and active transport, in 1D, 2D and 3D. In parallel we have developed with the Bolhuis group from the University of Amsterdam a multiscale algorithm, MD-GFRD, that combines the power of GFRD at mesoscopic length scales with the resolution of MD or BD at microscopic length scales. To learn more about GFRD, or to download the code, please visit www.GFRD.org.
Picture on the right shows a snapshot from a GFRD simulation.
A list of selected publications can be found here
FFS
Forward Flux Sampling (FFS) is one of the most widely used techniques to simulate rare events in both equilibrium and non-equilibrium systems. It has been applied to a very wide range of problems, from crystal nucleation, droplet formation, evaporation, nucleation in scale-free networks, DNA condensation, hydrophobicity, assembly in equilibrium colloidal systems and non-equilibrium active matter systems, phase separation in alloys, coalescence, wetting, shape transformations of vesicles, magnetic switches, protein folding and protein aggregation, polymer translocation, to genetic switches in cellular systems.
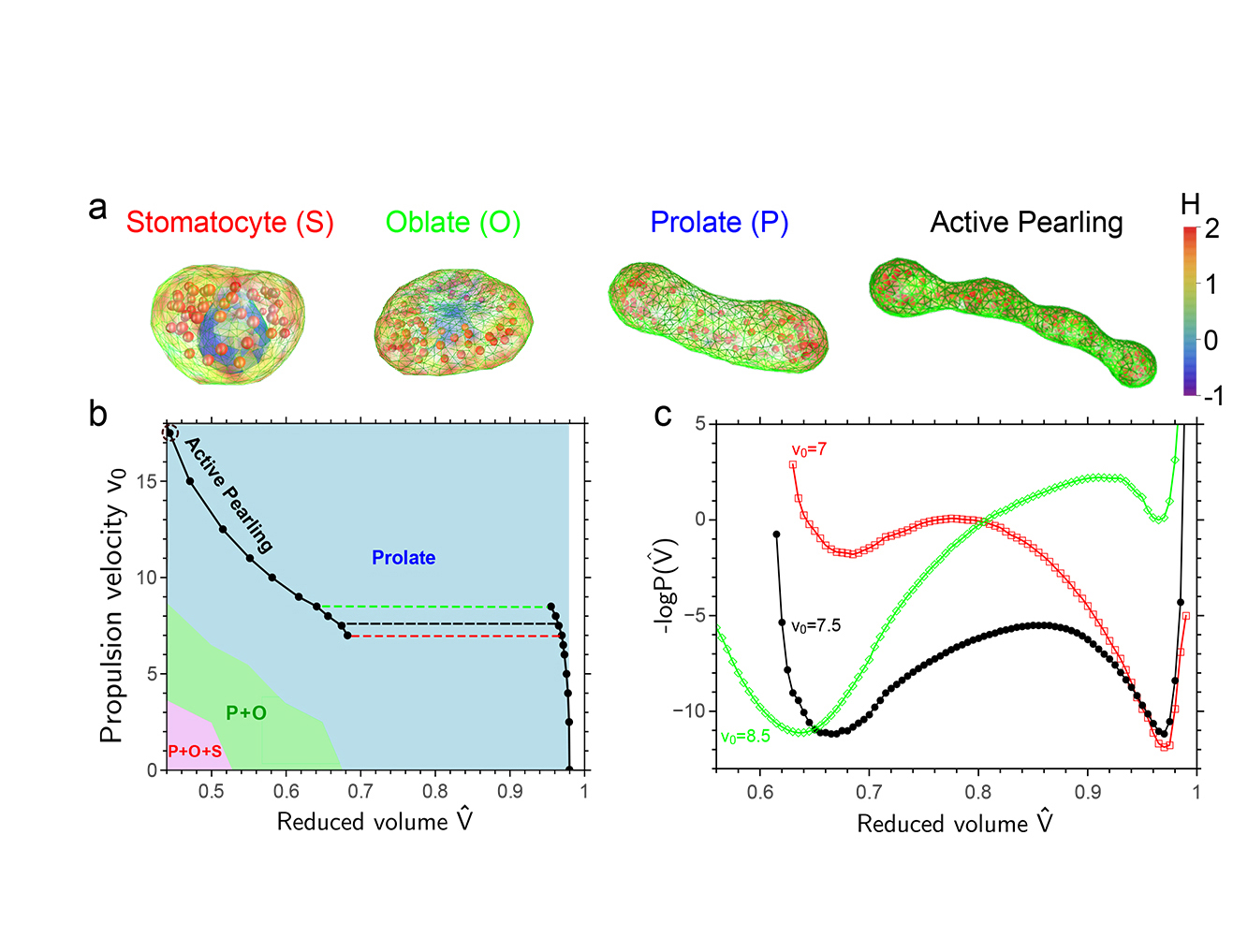
Picture to the right shows the phase diagram of shape transformations of vesicles that contain active particles, computed using FFS.
A list of selected publications can be found here