Studying individual molecules to understand how a cell’s inner life makes cells function
To understand the fundamental processes of life we need to understand the language that cells use to communicate with each other and to orchestrate their joint behavior. This is important for vital processes such as DNA replication, protein synthesis and signal transmission. Since cells communicate through interactions of molecules, researchers at AMOLF and the Max-Planck Institute of Biochemistry in Munich developed a technique to study individual molecules in model membranes. They first published the results in 2021 (see reference below). Using self-renewing DNA-based fluorescent labels, they were able to observe them for extended times. Overcoming the constraints of conventional fading fluorescent markers, the new technique enables unprecedented quantitative studies of molecular interactions in living systems. Now, their latest advancements on this DNA-PAINT based single-particle tracking method (DNA-PAINT-SPT) also enables them to see individual molecules binding to each other in live cells. This study was published in Nature Communications on July 19th (see reference below).
Self-renewing DNA-based fluorescent labels
The researchers developed a fluorescent label that continuously exchanges fluorophores. To establish this, they use a method called DNA-PAINT, in which fluorescently-labeled oligonucleotides reversibly bind to a single-stranded DNA handle attached to the target molecule. The researchers engineered sets of DNA strands that incorporate multiple binding sites for fluorescently-labeled oligonucleotides. They optimized the turnover kinetics and provided two sets of orthogonal sequences for dual-color labelling, an essential requirement for interaction studies. As a result, individual proteins in reconstituted systems and live cell membranes can now be tracked for several minutes with 20-30 nanometer precision, a leap forward in observational precision and temporal dynamic range.
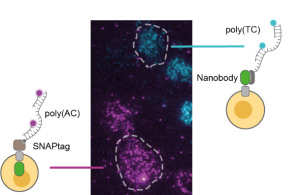
Control experiments
The researchers demonstrated that this improvement in performance allows the detailed quantification of two-dimensional binding constants of a model membrane protein. To rule out artifacts introduced by the DNA label, they conducted meticulous control experiments. These experiments involved measurements of the diffusion constants of proteins labelled with various methods and conducting additional controls with non-interacting proteins. The absence of these unwanted effects, combined with the straightforward one-to-one molecular targeting achievable through standard tagging methods, positions DNA-PAINT-SPT as a notably superior labelling approach. The new technique paves the way for more extensive studies of membrane protein interactions, a crucial yet challenging area of research.

References
Christian Niederauer, Chikim Nguyen, Miles Wang-Henderson, Johannes Stein, Sebastian Strauss, Alexander Cumberworth, Florian Stehr, Ralf Jungmann, Petra Schwille & Kristina A. Ganzinger, Dual-color DNA-PAINT single-particle tracking enables extended studies of membrane protein interactions, Nature Communications, 2023, https://doi.org/10.1038/s41467-023-40065-8
Florian Stehr, Johannes Stein, Julian Bauer, Christian Niederauer, Ralf Jungmann, Kristina Ganzinger & Petra Schwille, Tracking single particles for hours via continuous DNA-mediated fluorophore exchange, Nature Communications, 2021, https://doi.org/10.1038/s41467-021-24223-4


