How bacteria cope with randomness
Bacterial behavior is profoundly influenced by continuous random perturbances in their regulatory pathways. Researchers from institute AMOLF and Utrecht University found a new way by which bacteria cope with these perturbances. By performing microscopy measurements on individual bacteria, and mathematical modeling, they found that a regulatory mechanism well-studied for its response to external conditions also has a previously unrecognized role in dealing with metabolic fluctuations. The study yields new insights into cellular homeostasis and regulatory interactions. The results were published in the journal Cell Reports.
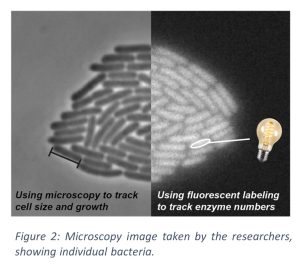
By combining experimental observations with genetic engineering and stochastic mathematical models, the work by Martijn Wehrens and his collaborator Laurens Krah from the group of Rutger Hermsen at Utrecht, shows how metabolic regulation not only performs its known role in choosing the appropriate amount of enzymes for a particular environment, but also responds to random fluctuations in internal metabolites. In the research field, it is well-known that these random fluctuations can produce real-world effects. For example, some bacteria swim much faster than others. Or, some individual bacteria reproduce faster than others under the same conditions. These fluctuations occur because fundamental molecular processes inside bacteria are partly based on chance, such that enzymatic and molecular concentrations continuously fluctuate randomly. To thrive in a particular environment, bacteria need to keep these random internal fluctuations in check. The dual function of the bacterial regulation revealed by the team of researchers allows bacteria to tame these fluctuations and keep their range of behaviors in check and more fine-tuned to their environment.
Reference
M. Wehrens, L.H.J. Krah, B.D. Towbin, R. Hermsen, and S.J. Tans, The interplay between metabolic stochasticity and cAMP-CRP regulation in single E. coli cells, Cell Reports, 42(10), 113284: 1-23, October(2023).
doi.org/10.1016/j.celrep.2023.113284


