Evolutionary conflicts predicted
Evolution seems to be an unpredictable process. However, predicting the constraints of evolution is possible. Researchers from AMOLF and the French ESPCI have demonstrated this using their mathematical method, followed by experiments on bacteria. On June 24, they publish their results in the journal Cell Systems.

Cells make use of close collaboration between proteins. Such regulatory networks enable cells to interpret their environment and therefore to switch ‘on’ the right gene, for example to express the enzyme that can break down antibiotics. This is vital for the survival of bacterial cells, and their regulatory networks are therefore under continuous evolutionary selection. However, it is difficult to predict how they will ultimately evolve.
The researchers opted for the following approach. They first made a new genetic network in the bacteria E. coli, enabling two environmental molecules (A and B) to influence the expression of a fluorescent protein. Instead of predicting the evolution of this network by examining random mutations, the researchers focused on all possible future functions of the network, and the constraints these entail. One of the questions they could pose was: ‘Can we predict whether evolution can ensure that the cells are only fluorescent if molecule A is present, and molecule B is not?’
To answer this question, the researchers developed a new mathematical method. Determining the evolutionary constraints of the genetic network proved to be simple using a graphical approach. This analysis indicated that the evolutionary goal described above was not easy to achieve. It would only be possible with a far-reaching evolutionary adaptation, for example in the detection of molecule A.
The researchers tested this prediction by allowing the bacteria to evolve in a controlled manner. They first allowed a large population of bacteria to randomly mutate, and subsequently placed these in a conical flask without A and B, and then with A but without B, etc. These experiments were set up in such a way that competition arose between the various mutants; the mutant which performed its task the best multiplied the most. The results revealed exactly what had been predicted: at the start of the evolution, the bacteria came close to the right solution, but not entirely. They responded either well to A, or well to B, instead of responding well to both signals. A sort of conflict arose between two evolutionary goals. After a longer period of evolution, it proved possible to solve the conflict – and a genetic analysis indeed revealed a far-reaching innovation of one of the proteins.
Being able to predict is a holy grail within evolution. In some disciplines, such as engineering or physics, making predictions is very normal, but within biology and even evolution this is far more difficult. Once again, many discussions are now taking place about the possibilities or impossibilities of making predictions and what this would require. Practical concepts and experiments such as these are beginning to provide an answer.
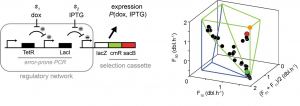
Schematic illustration of the network in E. coli (left), and the result of the evolution experiments and predictions (right). The network detects two molecules Dox and IPTG, and as a response allows a set of genes (lacZ, cmR, and sacB) to be expressed. The network is mutated by means of a PCR reaction, and the set of genes ensures the natural selection. The three axes that can be seen on the right show the growth rate in three different directions. The blue and green lines show the predicted evolutionary constraints. The points are the evolved E. coli strains, starting with the green point and ending at the red point. The intermediary black points follow the predicted constraints.
Reference
Manjunatha Kogenaru, Philippe Nghe, Frank J. Poelwijk, Sander J. Tans, Predicting Evolutionary constraints by identifying conflicting demands in regulatory networks, Cell Systems 10, 1-9, June 24 (2020).


