A magic top hat for protein folding
“Folding acceleration mechanisms have never been shown before for any protein system. This is arguably the most beautiful protein research we have ever done in our laboratory! A highly surprising mechanism, and all observations and control measurements match perfectly,” Tans said.
How does a cavity fold a protein?
Proper folding is crucial for all proteins. Their essential tasks in cells depend on their threedimensional shape. However, forming the correct shape is far from obvious. Following the synthesis of a long chain of amino acids, the chain must fold. In this process, all amino acid residues must find the right neighbors to create the correct overall shape. This process can easily go wrong, with different chains sticking to each other before they are folded, and hence forming harmful protein aggregates.
A class of helper proteins called chaperones assist with protein folding, but a key issue remains poorly understood: do chaperones only prevent aggregation or also accelerate the folding process itself, and if so, how?
To study this question, Tans and his team member Mohsin Naqvi focused on the chaperone GroEL, together with their collaborators at Texas A&M University. The GroEL chaperone has a cavity that can be covered and closed by its partner chaperone GroES. This remarkable shape has fascinated biologists for a long time, and led to suggestions that amino acid chains are folded by compression within this closed cavity. Another proposed mechanism involved unfolding of misfolded proteins. But what type of mechanism can indeed accelerate folding has remained controversial.
Single molecule technique
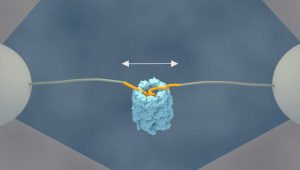
The researchers at AMOLF have now demonstrated a mechanism that accelerates protein folding. Tans explaines: “Using a single molecule technique, we have been able for the first time to follow single protein chains as they enter the GroEL cavity. What we saw was very striking; the unfolded chain was forcefully being pulled into the GroEL chamber, where it collapsed on top of itself. Next, we saw it almost instantly snap into the proper structure, even those proteins that normally fold very slowly.”
For these experiments, Tans and his colleagues used optical tweezers to attach the two ends of an amino acid chain to two micron-sized beads, which can be moved around by laser beams. In this way they can manipulate and measure the distance between the protein ends.
When they added the GroEL chaperone to the solution, they observed that the two ends moved closer together. First gradually, the protein chain started collapsing inside the GroEL cavity. Then rapidly, almost all of the amino acids were folding into their proper position.
Intuitive explanation
“It is wonderful to actually see this happening,” said Tans. “This mechanism also provides an intuitive explanation for the folding acceleration, because amino acids must of course first come close together in order to fold. It is well known that amino acid chains can collapse by themselves. We show for the first time that this collapse is strengthened and regulated in the cell by chaperones.”
Tans continues: ”Our findings show that the internal structure, energy and dynamics of collapsed states can be modified by another protein, the chaperone, and hence can be subjected to regulation pathways.”
Perfect control experiments
In addition to the main observation of the protein ends being pulled together by GroEL, the researchers report an exhaustive study that includes diverse conditions, mutants, protein substrates, and assay types.
In one control experiment, the researchers removed the so-called C-terminal tails of GroEL, small extensions inside the cavity of GroEL. After truncating these tails, the folding probability of the protein decreased, showing that the collapse and folding takes place inside the GroEL cavity.
In another control experiment, the researchers focused on GroES, the ‘lid’ on the cavity of GroEL. They found GroES was not required to accelerate the slowest folding transition, though it was critical to create fully formed proteins and eject them from the GroEL cavity. Tans explaines: “This shows that the discovered mechanism is different from steric confinement. If it were steric confinement, protein folding would not be accelerated without the lid.”
Lastly, the researchers attached a fluorescent label to GroEL. Using a scanning laser beam to create an image, GroEL showed up exactly between the two beads to which the protein is attached, after which folding took place, showing directly that the folding protein was indeed bound to GroEL.
Striking observation
Tans reflects that regulating the collapse of proteins via this newly discovered mechanism is likely a more general chaperone strategy. “We conjecture that our collapse modulation mechanism may be exploited throughout the protein quality control machinery, including by the human homologue TRiC. But, it could also play a role in controlling protein phase separation, where the hydrophobic collapse is also important.”
The notion that protein collapse and folding speeds can be regulated may shine a new light on diseases where protein folding and aggregation plays an important role. For instance, it offers new avenues to study the regulation of disordered oncogenic c-Myc/Max complex, the phase separated FUS proteins, which play a role in ALS and the aggregating tau protein associated with degenerative brain diseases.
Reference
Mohsin M. Naqvi, Mario J. Avellaneda, Andrew Roth, Eline J. Koers, Antoine Roland, Vanda Sunderlikova, Gunter Kramer, Hays S. Rye and Sander J. Tans
Direct observation of GroEL-stimulated protein collapse and folding, Science Advances, 8, 4 March (2022).
DOI: 10.1126/sciadv.abl6293


