New algorithm improves ability to track movements of cells in organs
Frustrated by the time cost of manually checking cell tracking data, AMOLF PhD student Max Betjes developed an algorithm to more efficient cell-tracking analysis. He validated this automated analysis on cell movements in miniature organs, and the results are now published in Nature Methods.
3D cell tracking and the problem with AI
3D cell tracking is a method used to look at the development of cells in organs, both during normal growth and when they are affected by disease. Computer algorithms based on neural networks can look at a series of microscope images and tell you where and how the cells are moving.
Such analysis has been limited by the lack of error indication in these types of automated processes, which would tell you how confident you can be in the results. “This is a general problem with AI,” says AMOLF PhD student Max Betjes; “it can give a prediction, but often no indication of whether it knows for sure.”
Because of this ‘black box’ processing, the outputs of cell-tracking algorithms need to be manually checked by the user. But this is cumbersome, and still doesn’t give a precise indication of how correct the data is.
As frequent users of cell-tracking programs for their own research, Max and his team member Rutger Kok, who now works at the UMC Utrecht, decided to try to address this issue.
Organoids, blastocysts and … worms?
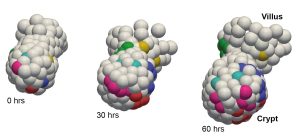
Max solved this problem by modifying the AI analysis to be much stricter with statistics. “We forced the AI parts of the algorithm to state their predictions in terms of probabilities,” explains Max, “and with those probabilities we can calculate the expected error rate.”
This gave him an algorithm that outputs both tracks and an error indicator in a fully automated process. Users can then correct low-certainty parts of the data or automatically filter these out. To prove that it works, he used pre-validated data from their own lab. This showed that his algorithm works for cells in intestinal organoids, which are clusters of cells that behave like an intestine, and whose cells are notoriously hard to track. “It would have been hard to develop something like this in a vacuum,” says Max, “which is why it was valuable that we could test it on microscopy data that we actually use for research.”
In addition to his lab’s own data, Max tested the algorithm on publicly available cell data with cells from mouse blastocysts (very early-stage embryos), and C. elegans worms (1-millimeter nematodes that live in soil). “Publicly available datasets are fun,” says Max. “The C. elegans data was part of something called the ‘Cell Tracking Challenge,’ where they rank your data analysis.” Again he was able to validate the reliability of his algorithm, with his automated C. elegans analysis now ranking in first place in the Cell Tracking Challenge.
Expanding possibilities for organoid research
A big motivation for studying organoids is their potential for use as living models for real organs in research on drugs and diseases. “But if you’re manually checking cell tracks,” says Max, “you can’t do more than a few organoids.” Automating the cell tracking process is one step in being able to apply organoid research on a larger, more clinical scale.
However, Max emphasizes that to achieve this a lot more work must be done. “Accurate cell tracking is not the only ingredient needed,” he explains. One challenge of applying this to medical research is imaging many samples with consistently high resolution. “More publicly available training data could make the tracking more robust,” says Max.
At the same time, this development could draw more interest in cell tracking research for its potential in larger-scale clinical studies. With further research and collaboration, Max’s cell tracking software can be bolstered for wider use.
Try it yourself
Max and his team within the groups of Sander Tans and Jeroen van Zon want to invite potential collaborators to try the software with their own data, which will help them develop it further. They’ve created a website for this, where you can also run part of the program using example data.
Join the collaboration, or try the software with example data here: OrganoidTracker.org
Publication
Max A. Betjes, Rutger N. U. Kok, Sander J. Tans & Jeroen S. van Zon. Cell tracking with accurate error prediction. Nature Methods (2025). https://doi.org/10.1038/s41592-025-02845-6


