Biochemical systems
3D tracking of micron-sized particles
Group: Systems Biology
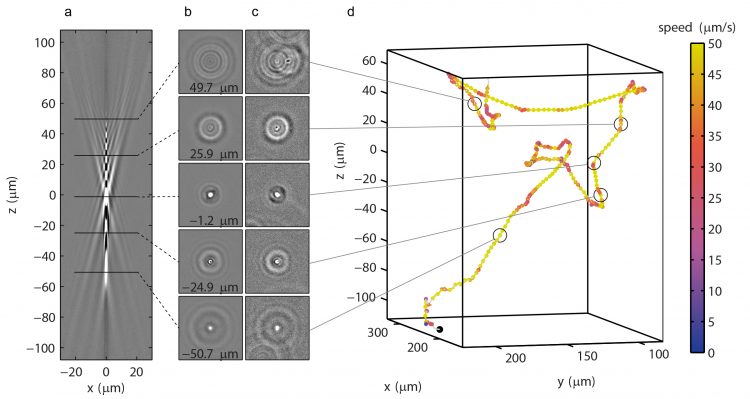
We have developed a simple, robust, and broadly applicable, high-throughput 3D tracking method based on computational image analysis, which requires only a standard phase contrast microscope, a camera and a PC.
Potential applications
- Tracking 3D movements of colloidal particles, micro-organisms, and characterizing microfluidic flow patterns
Reference
K. Taute, S. Gude, S.J. Tans and T.S. Shimizu, High-throughput 3D tracking of bacteria on a standard phase contrast microscope, Nature Commun. 6, Article number: 8776: 1-9 (2015)
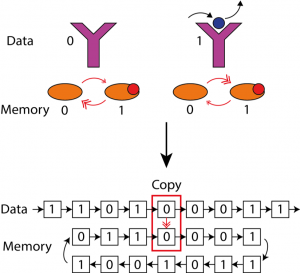
Biochemical computing and cellular information transmission
Group: Biochemical Networks
In our group we combine ideas from statistical physics and stochastic thermodynamics to study biochemical computing and cellular information transmission, using analytical theory and novel computer simulation techniques.
Potential applications
- Biochemical computing
Reference
T.E. Ouldridge, C.C. Govern and P.R. ten Wolde, Thermodynamics of Computational Copying in Biochemical Systems, Phys. Rev. X 7, 2, Article number: 021004: 1-13 (2017)
Developmental biology, nematode worms, organoids, microscopy, image analysis and machine learning
Group: Quantitative Developmental Biology
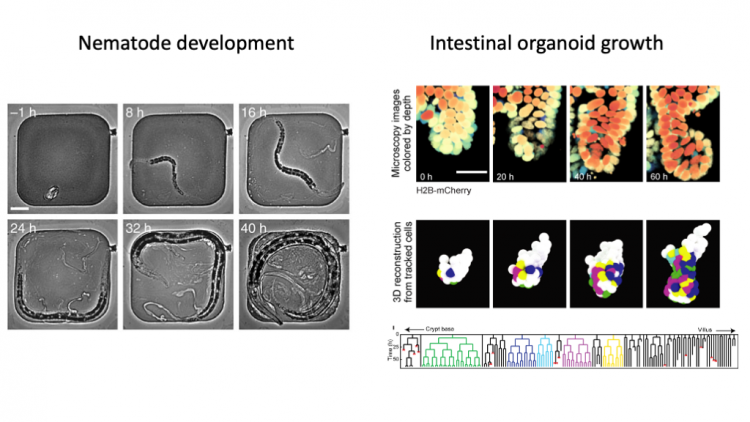
We use advanced microscopy, quantitative image analysis and mathematical modelling, to understand how cells make decisions during development, how timing of development is controlled and complex tissues, like the human intestine, renew themselves.
Potential applications
- Drug screening
- In vitro organ models
- Plant and human diseases of parasitic nematodes
Reference
N. Gritti, S. Kienle, O. Filina and J.S. van Zon, Long-term time-lapse microscopy of C. elegans post-embryonic development, Nature Commun. 7, Article number: 12500: 1-5 (2016)
Particle-based simulations of diffusion in complex geometries and mixed boundary conditions
Group: Theory of Biomolecular Matter
We develop state-of-the-art stochastic approaches to solving diffusion problems involving non-trivial geometries (curved surfaces, diffusion bottlenecks, …) with mixed Robin type boundary conditions, which do not require spatial discretisation.
Potential applications
- reaction diffusion systems
- thermal design

Cellular dynamics in organoids, protein synthesis and folding
Group: Biophysics
At the molecular level, we use optical tweezers and single-molecule fluorescence to study how chaperones fold amino-acid chains into functional proteins. At the cellular level, we use time-lapse microscopy and image analysis to understand how organoids self-organise.
Potential applications
- 3D microscopy techniques
- Machine learning enabled microscopy analysis
- Single-molecule methods
Reference
M.J. Avellaneda, K.B. Franke, V. Sunderlikova, B. Bukau, A. Mogk and S.J. Tans, Processive extrusion of polypeptide loops by a Hsp100 disaggregase, Nature 578, 317-320 (2020)


