Imaging individual and collective bacterial motility
Imaging motile behavior in microfabricated environments
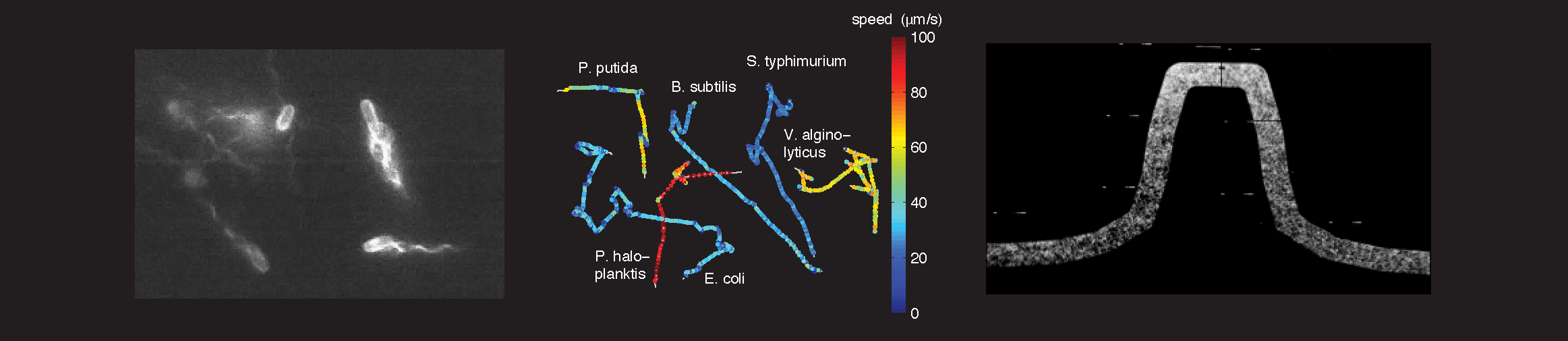
One of the major difficulties in understanding intracellular networks stems from uncertainties about the environments in which cells function. We develop microfluidic devices1 and new imaging techniques2 to quantify the behavior of bacteria in controlled microenvironments 3. Soft lithography techniques are used to obtain transparent silicone-rubber (PDMS) chips with micron-scale indentations that serve as fluid channels when mated with an opposing surface. Such devices are well suited for precise control of the cells’ fluidic environment, since turbulent flows are non-existent at these small scales.
References
- Ahmed, T., Shimizu, T. S. & Stocker, R. Bacterial chemotaxis in linear and nonlinear steady microfluidic gradients. Nano Letters 10:3379-3385 (2010)
- Taute,K.M., Gude,S., Tans, S.J. & Shimizu, T.S. High-throughput 3D tracking of bacteria on a standard phase contrast microscope, Nature Commun 6, 8776: 1-9 (2015)
- Lazova, M. D., Ahmed, T., Bellomo, D., Stocker, R. & Shimizu, T. S. (2011). Response rescaling in bacterial chemotaxis. PNAS 108:33870-33875.