Single-cell FRET measurements of signaling diversity and temporal variability
We have developed a FRET microscopy method that allows measurements of protein signaling dynamics in individual bacteria1.
We want to understand the design of intracellular signaling networks, both in terms of mechanism and function. For this purpose, it is crucial to characterize the dynamic response of the network to input stimuli. The pathway used by the bacterium Escherichia coli to sense chemical gradients is an excellent example of a simple signaling circuit (Fig. 1) that performs a well-defined function.
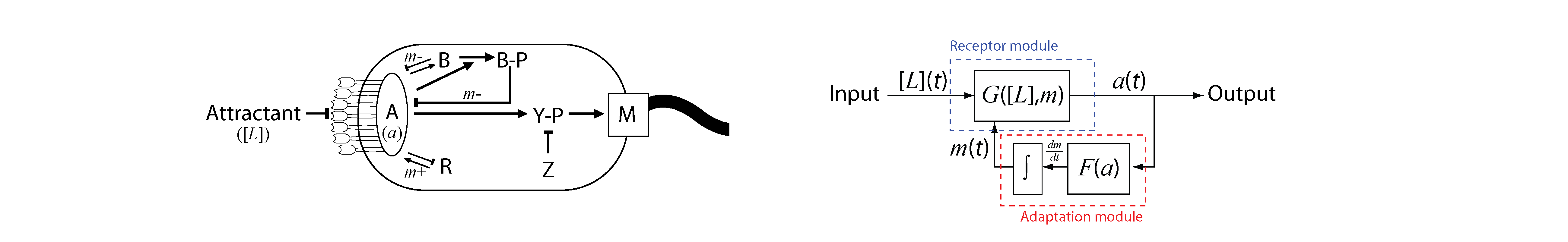
Fig. 1. The E. coli chemotaxis signaling pathway. (Left) Biochemical view, illustrating interactions between protein components (receptors, A, R, B, Y, Z, M). (Right) Coarse-grained view, in which the three essential dynamical variables (ligand in put, [L](t), kinase activity, a(t), covalent modification levels of receptors, m(t)) are linked by two “transfer functions” (F(a), G([L],m)) representing the combined action of multiple protein species.
In vivo FRET measurements of signaling dynamics
We use Förster resonance energy transfer (FRET) to study the dynamic input-output relations of signaling in living cells. Responses to time-varying stimuli applied in microfluidic chambers reveal the essential input/output relationships of the system, to reveal the “shape” of the essential transfer functions (Fig. 2). These measurements form the basis for studying both their functional implications, as well as their mechanistic implementation at the molecular level2,3,4.
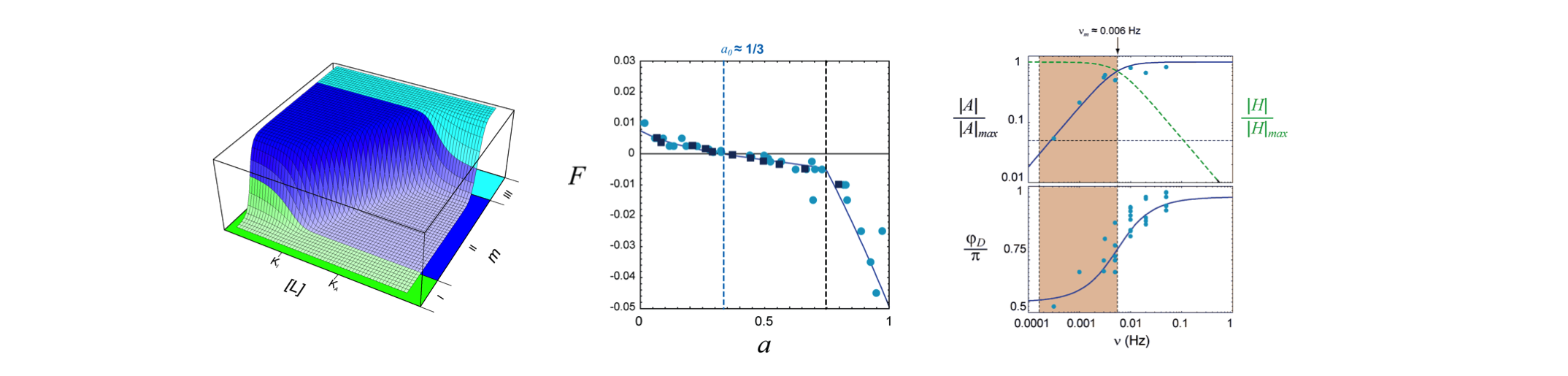
Fig. 2. Transfer functions of E. coli chemotaxis signaling. (Left) The receptor-module transfer function, G([L],m), relates ligand input, [L], and methylation feedback, m, to the kinase activity, a(t). (Center) The adaptation-module transfer function, F(a), determines the rate of change of the feedback signal, dm/dt, given the current kinase activity, thereby providing integral feedback. (Right) The frequency response of the pathway measured by FRET (blue points) closely match model predictions based on the measured transfer functions F(a) and G([L],m).
FRET in single bacteria reveals signaling variability
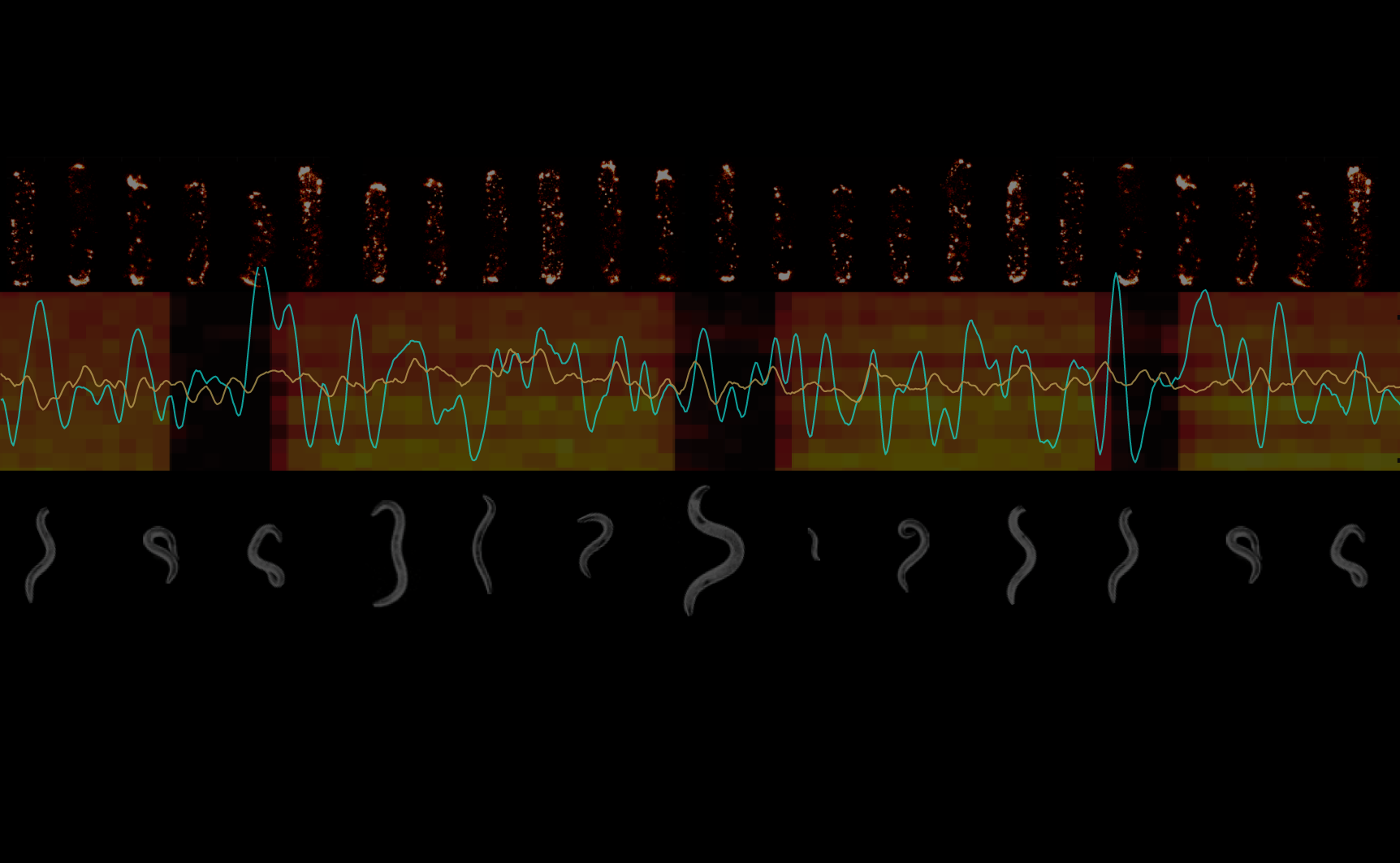
Wile FRET has been a powerful tool for investigating signaling dynamics in vivo, the technique is applied to bacteria mostly at the population level, due to the small size of cells and relatively low copy number of labeled proteins. We have extended in vivo FRET to the single-cell level, to enable measurement of signaling dynamics in individual cells (Fig. 3). These measurements reveal pervasive variability in signaling, between cells in a population, and within single cells across time1. Such data strongly constrain mechanistic models of signaling2,5, and provide new insights regarding the functional consequences of molecular noise6,7.
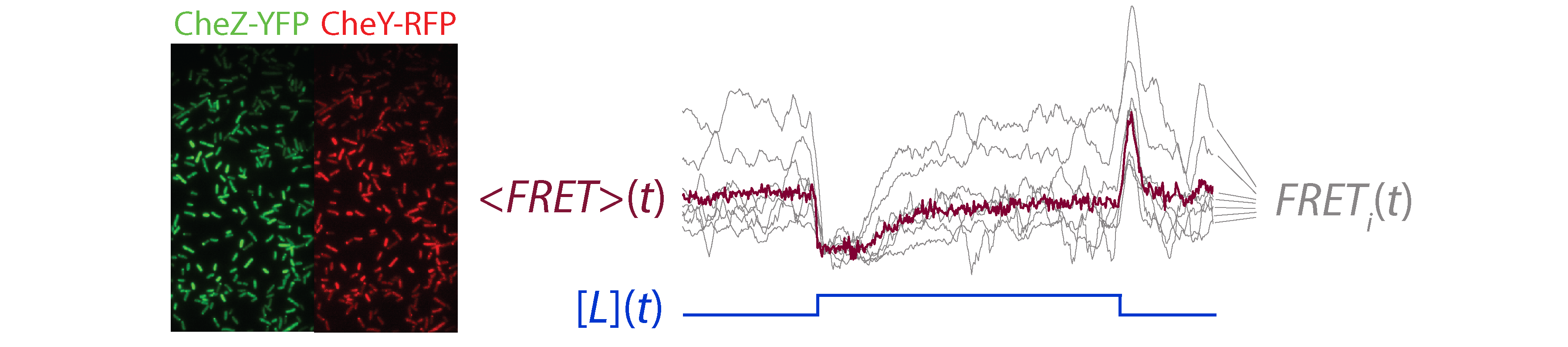
Fig. 3. Single-cell FRET measurements of signaling dynamics. (Left) Movies of FRET donor (CheZ-YFP) and acceptor (CheY-RFP) signals are acquired simultaneously and segmented to compute single-cell FRET signals. (Right) FRET response time series of individual cells from an experiment in which all cells were simultaneously stimulated with a step change in the ligand signal, [L](t). The single-cell responses, FRETi(t), demonstrate substantial differences from one another, as well as the population averaged response, <FRET(t)>.
References
- Kamino, K., Keegstra, J.M., Long, J., Emonet, T. & Shimizu, T.S. (2020). Adaptive tuning of cell sensory diversity without changes in gene expression. (submitted).
- Keegstra, J.M., Kamino, K., Anquez, F., Lazova, M.D., Emonet, T. & Shimizu, T. S. Phenotypic diversity and temporal variability in a bacterial signaling network revealed by single-cell FRET. eLife 6:e27455 (2017)
- Tu, Y., Shimizu, T. S. & Berg, H. C. Modeling the chemotactic response of Escherichia coli to time-varying stimuli. PNAS 105, 14855-60 (2008)
- Shimizu, T. S., Tu, Y. & Berg, H. C. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol Syst Biol 6:382 (2010)
- Lazova, M. D., Ahmed, T., Bellomo, D., Stocker, R. & Shimizu, T. S. Response rescaling in bacterial chemotaxis. PNAS 108:33870-33875 (2011)
- Shimizu, T. S., Aksenov, S. V. & Bray, D. A spatially extended stochastic model of the bacterial chemotaxis signalling pathway. J Mol Biol 329, 291-309 (2003)
- Korobkova, E., Emonet, T., Vilar, J. M., Shimizu, T. S. & Cluzel, P. From molecular noise to behavioural variability in a single bacterium. Nature 428:574-578 (2004)
- Flores, M., Shimizu, T. S., ten Wolde, P. R. & Tostevin, F. Signalling noise enhances chemotactic drift of E. coli. Phys Rev Lett 109:148101 (2012)