A new spin on making minimal cells
The ability of a cell to separate its own matter from its surroundings is a basic requirement for life. A team of researchers at AMOLF and Delft University of Technology have managed to create a synthetic container, or lipid vesicle, that is able to hold a range of different biological systems: from a cytoskeleton to entire E.coli bacteria. Their findings on this optimized cDICE method, which has the potential to reveal the inner workings of life, are published in ACS Synthetic Biology on June 29, 2021.
Lipid vesicles are an ideal candidate to separate the inside of the cell from its outside surroundings, which is central to building a minimal cell. However, controlling a vesicle’s inner content has so far proven tricky. The researchers demonstrate how a comparatively simple method, which is based on an earlier method called cDICE, can lead to vesicles with a range of minimal systems inside. In this method, vesicles are formed by driving capillary-generated water-in-oil droplets across an oil-water interface using centrifugal force, created by spinning a rotating chamber.
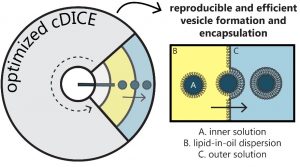
The researchers found that tightly controlling environmental conditions while using this method, in particular air humidity, was crucial to reproducibly form many high-quality vesicles, both for months of experiments and across different labs. Furthermore, they found that by tuning the composition of the oil phase, molecular components could more efficiently be enclosed inside these vesicles.
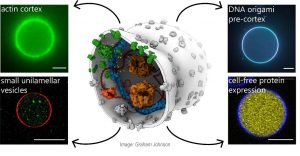
Together, these changes allowed the scientists to make vesicles that can contain a range of minimal systems inside: for example a minimal (actin) cytoskeleton, membrane-anchored DNA nanostructures, but also small unilamellar vesicles to mimic organelles, and a functional protein synthesis machinery (PURE; protein synthesis using recombinant elements). This shows that the optimized cDICE method can lead to vesicles with a range of minimal systems that are all required for the generation of a minimal cell.
By publishing these findings alongside the detailed protocols and the blueprints for the cDICE device, the researchers hope that their optimized cDICE method can easily be picked up by others and become a standard method in synthetic biology and other fields such as biophysics. This project is part of the larger project ‘BaSyC: Building a Synthetic Cell’, in which researchers across the Netherlands have teamed up to build a minimal cell. On a European level, researchers and institutes are working towards the same goal with the European Synthetic Cell Initiative.
Collaborating groups: Physics of Cellular Interaction Group (AMOLF), Koenderink Lab, Cees Dekker Lab and Danelon Lab (all TU Delft, department of Bionanoscience).
Reference
Lori van de Cauter, Federico Fanalista, Lennard van Buren, Nicola De Franceschi, Elisa Godino, Sharon Bouw, Christophe Danelon, Cees Dekker, Gijsje H. Koenderink and Kristina A. Ganzinger, Optimized cDICE for Efficient Reconstitution of Biological Systems in Giant Unilamellar Vesicles, ACS Synth.Biol 2021: https://doi.org/10.1021/acssynbio.1c00068.


